Page History
...
The MetaBiobank service offers the ability to import external sample collections from files. If you have a CSV file with information concerning individual samples (one sample per row), you can use the automated import facility to import it into one of your existing biobanks. This is done by clicking the Upload samples tab in the biobank management panel.
The service will present you with a page where you need to specify the name of the collection your samples will be uploaded to (a new collection will be created automatically if the selected collection doesn't yet exist) and choose a file stored on your hard drive to read samples from.
Below we present a sample (simplified) data import file:
Kod,Opiekun,Kod pacjenta,Płeć,Wiek,Materiał,Temperatura,Pobrane dane
M150004247,JSMITH,p031,Female,25,DNA,-20,4/30/2015 13:07
M150004248,JSMITH,p032,Female,30,DNA,-20,4/30/2015 16:55
In this case the file contains a header row followed by a set of data rows, each of which contains a description of an individual sample.
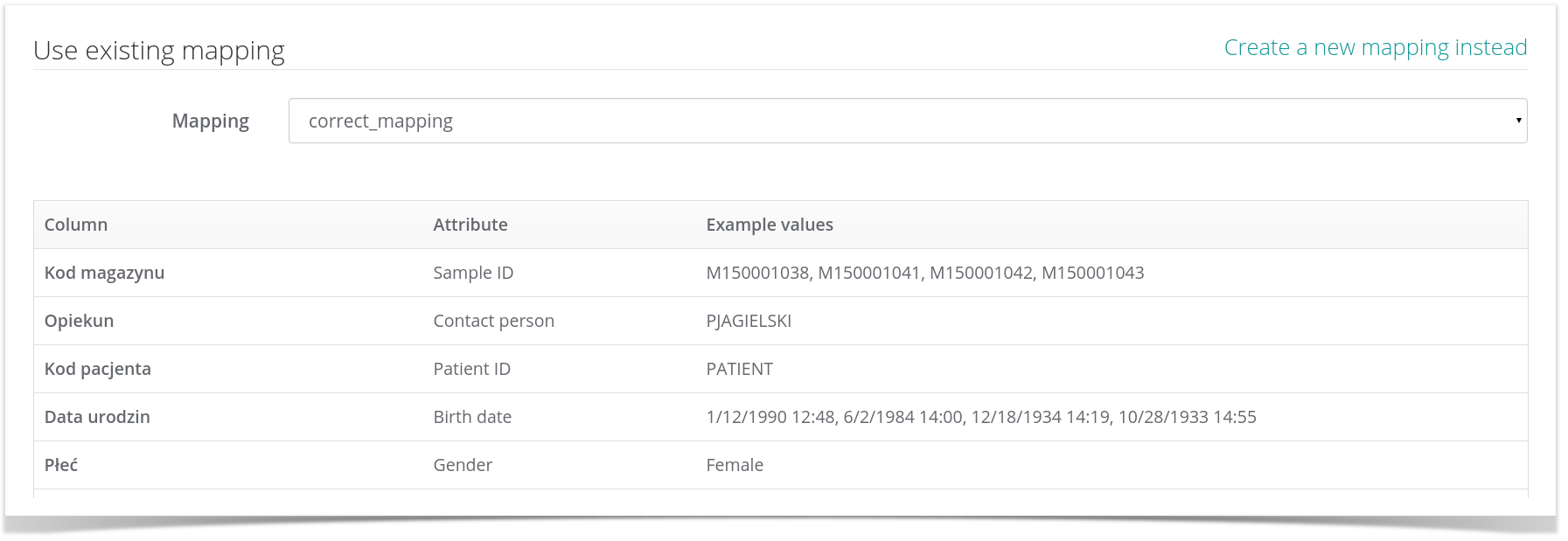
We are aware We are aware of the fact that not exports are created equal and may include different types of data regarding samples (expressed as individual columns in your CSV file). In order to enable your data to be imported in a meaningful fashion, MetaBiobank asks you - as the second step in the import process - to specify a mapping between CSV columns and metadata terms specified by the MIABIS biobank description standard. Clicking Upload will validate your file and take you to the next step in the import process where you can either specify an existing mapping, as shown in the following image:
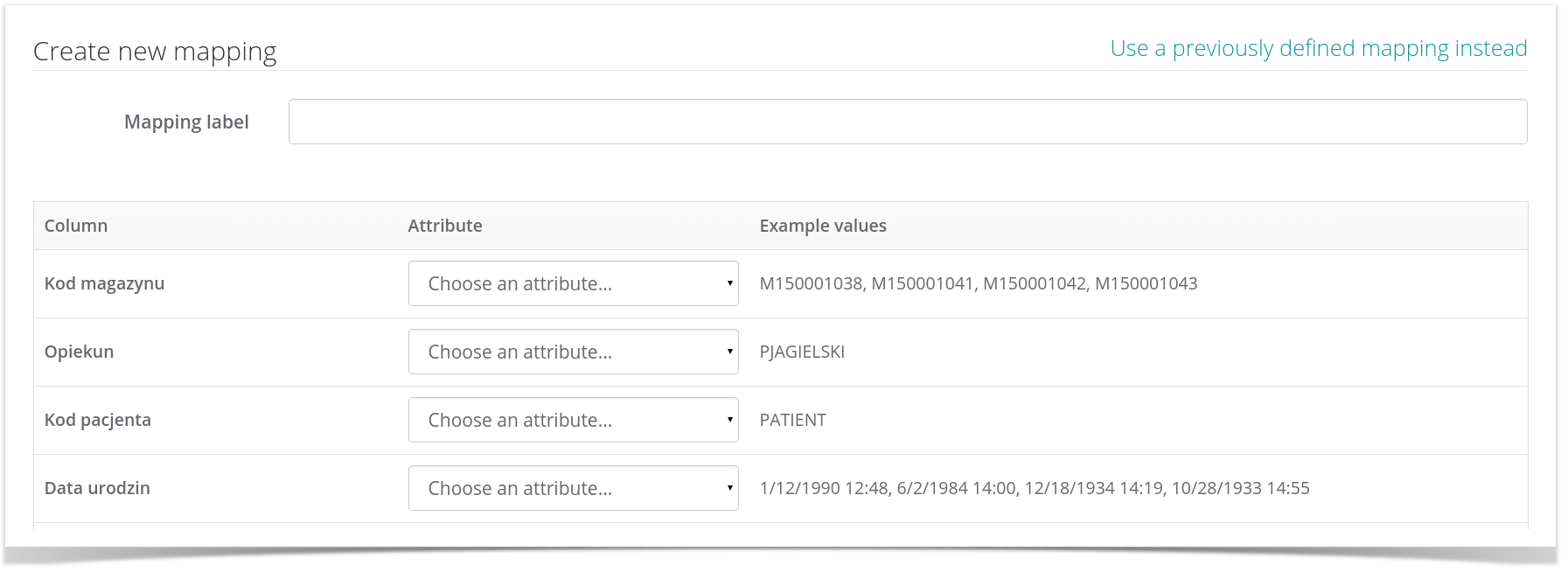
...or, alternatively, create and save a new mapping for later use by clicking the Create a new mapping instead option:
a new one. A mapping is a collection of column/term assignments where column is one of the data columns in your CSV file while term is one of the available MIABIS terms. For example, if you assign column named 'Data pobrania' to the MIABIS term Sampled date, the MetaBiobank service will assume that all values of this column in the CSV file indicate the dates on which each sample was taken.
Note that while MetaBiobank will do its best to analyze and properly parse the data present in your CSV file, we cannot guarantee that a sensible match for a meaningful MIABIS value can be made in each case (for example - the date field may be blank or formatted in a way which the MetaBiobank service does not understand). In all such cases the 'problematic' data items will be copied to a separate field in the sample description. We have named this field Comments and it can later be used to retrieve data which the data import facility could not appropriately parse.
Also note that in order to be usable, a mapping must, at the very least, include mapping items for the following MIABIS terms:
- Sample ID - column which contains a unique identifier of each sample
- Material type - column which lists the type of biological material represented by each sample
- Storage temperature - column which lists the temperature at which each sample was stored.
Failure to specify mappings for any of these fields will make the mapping unusable until such time as the missing data is provided.